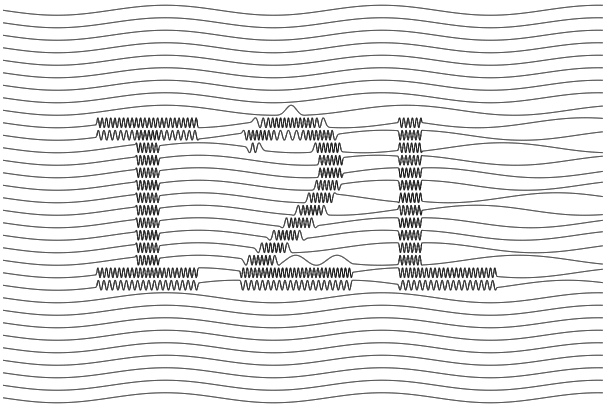
img2laser is a browser-based tool written entirely in Rust that can convert a bitmap image into a laser-ready SVG image using the sine wave line shading technique. In my original blog post, I showcased a Python notebook that performed the same task, but it was lacking in portability. I wanted to make the tool more […]